Tucked within its double-helix structure, DNA contains the chemical blueprint that guides all the processes that take place within the cell and are essential for life. Therefore, repairing damage and maintaining the integrity of its DNA is one of the cell’s highest priorities.
Researchers at Vanderbilt University, Pennsylvania State University, and the University of Pittsburgh, utilizing the U.S. Department of Energy Office of Science’s Advanced Photon Source (APS) at Argonne, discovered a fundamentally new way that DNA-repair enzymes detect and fix damage to the chemical bases that form the letters in the genetic code. Their discovery was reported in the journal Nature.
“There is a general belief that DNA is ‘rock solid’ – extremely stable,” said Brandt Eichman, associate professor of Biological Sciences at Vanderbilt, who directed the project. “Actually DNA is highly reactive and can be chemically modified a number of different ways.”
On a good day about 1 million bases in the DNA in a human cell are damaged. These lesions are caused by a combination of normal chemical activity within the cell and exposure to radiation and toxins coming from environmental sources including cigarette smoke, grilled foods, and industrial wastes.
“Understanding protein-DNA interactions at the atomic level is important because it provides a clear starting point for designing drugs that enhance or disrupt these interactions in very specific ways,” said Eichman. “So it could lead to improved treatments for a variety of diseases, including cancer.”
The newly discovered mechanism detects and repairs a common form of DNA damage called alkylation. A number of environmental toxins and chemotherapy drugs are alkylation agents that can attack DNA. When a DNA base becomes alkylated, it forms a lesion that distorts the shape of the molecule enough to prevent successful replication. If the lesion occurs within a gene, the gene may stop functioning. To make matters worse, there are dozens of different types of alkylated DNA bases, each of which has a different effect on replication.
One method to repair such damage that all organisms have evolved is called “base excision repair” (BER). In BER, special enzymes known as DNA glycosylases travel down the DNA molecule scanning for these lesions. When they encounter one, they break the base pair bond and flip the deformed base out of the DNA double helix. The enzyme contains a specially shaped pocket that holds the deformed base in place while detaching it without damaging the backbone. This leaves a gap (called an “abasic site”) in the DNA that is repaired by another set of enzymes.
Human cells contain a single glycosylase, AAG, that repairs alkylated bases. It is specialized to detect and delete ethenoadenine bases, which have been deformed by combining with highly reactive, oxidized lipids in the body. However, AAG also handles many other forms of akylation damage. Many bacteria have several types of glycosylases that handle different types of damage.
“It’s hard to figure out how glycosylases recognize different types of alkylation damage from studying AAG since it recognizes so many,” said Eichman. “So we have been studying bacterial glycosylases to get additional insights into the detection and repair process.”
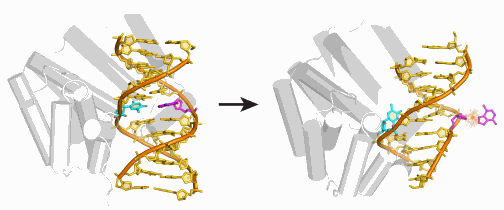
That is how the research team, using high-energy x-ray beams to carry out protein crystallography studies at the Life Sciences Collaborative Access Team 21-ID-D beamline at the APS, discovered the bacterial glycosylase AlkD with its unique detection and deletion scheme. All the known glycosylases work in basically the same fashion: They flip out the deformed base and hold it in a special pocket while they excise it.
AlkD, by contrast, forces both the deformed base and the base it is paired with to flip to the outside of the double helix. This appears to work because the enzyme only operates on deformed bases that have picked up an excess positive charge, making these bases very unstable. If left alone, the deformed base will detach spontaneously. But AlkD speeds up the process by about 100 times.
Eichman speculates that the enzyme might also remain at the location and attract additional repair enzymes to the site.
See: Emily H. Rubinson1, A. S. Prakasha Gowda2, Thomas E. Spratt2, Barry Gold3, and Brandt F. Eichman1, “An unprecedented nucleic acid capture mechanism for excision of DNA damage,” Nature 468, 406 (18 November 2010). DOI:10.1038/nature09428
Author affiliations: 1Vanderbilt University, 2Pennsylvania State University College of Medicine, 3University of Pittsburgh
Correspondence: [email protected]
Use of LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor. This research was supported by a grant fromthe American Cancer Society (to B.F.E.) and the NIH (RO1 CA29088 to B.G.). E.H.R. was supported in part by the Vanderbilt Training Program in Molecular Toxicology. Additional support for local crystallography facilities was provided by the Vanderbilt Center in Molecular Toxicology and the Vanderbilt-Ingram Cancer Center.
The original article by David F. Salisbury can be found in “exploration,” Vanderbilt University’s online research magazine: http://www.exploration.vanderbilt.edu/
Note: In other research at the APS (also at LS-CAT, along with other beamlines) researchers from the University of Chicago, the University of Wisconsin-Madison, and the Chinese Academy of Sciences observed, for the first time, an intermediate stage in the chemical process that repairs DNA methylation damage and regulates many important biological functions that impact health conditions such as obesity, cancer, and diabetes.
Work at Argonne and use of the Advanced Photon Source is supported by the U.S. Department of Energy Office of Science, Office of Basic Energy Sciences (DOE-BES), under Contract No. DE-AC-02-06CH11357. M. A. L.-M. acknowledges the Spanish MEC for a postdoctoral grant. M. v.V. was supported by the DOE-BES Office of Basic Energy Sciences, Division of Materials Sciences and Engineering, under Grant No. DE-FG02-03ER46097. S. C. and G. C. were supported by the U.S. National Science Foundation through Grants No. DMR-0552267 and No. DMR-0856234.
The Advanced Photon Source is an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory. The APS is the source of the Western Hemisphere’s brightest high-energy x-ray beams for research in virtually every scientific discipline. More than 3,500 scientists representing universities, industry, and academic institutions from every U.S. state and several foreign nations visit the APS each year to carry out applied and basic research in support of the DOE mission to understand, predict, and ultimately control matter and energy at the electronic, atomic, and molecular levels in order to provide the foundations for new energy technologies and to support DOE missions in energy, environment, and national security. To learn more about DOE x-ray user facilities.
Argonne National Laboratory seeks solutions to pressing national problems in science and technology. The nation's first national laboratory, Argonne conducts leading-edge basic and applied scientific research in virtually every scientific discipline. Argonne researchers work closely with researchers from hundreds of companies, universities, and federal, state and municipal agencies to help them solve their specific problems, advance America's scientific leadership and prepare the nation for a better future. With employees from more than 60 nations, Argonne is managed by UChicago Argonne, LLC for the U.S. Department of Energy's Office of Science.