The original MIT press release by Anne Trafton can be read here.
Industrial and agricultural activities produce large amounts of methane, a greenhouse gas that contributes to global warming. Many bacteria also produce methane as a byproduct of their metabolism. Some of this naturally released methane comes from the ocean, a phenomenon that has long puzzled scientists because there are no known methane-producing organisms living near the ocean’s surface. A team of researchers from MIT and the University of Illinois at Urbana-Champaign using the U.S. Department of Energy’s Advanced Photon Source (APS) has made a discovery that could help to answer this “ocean methane paradox.”
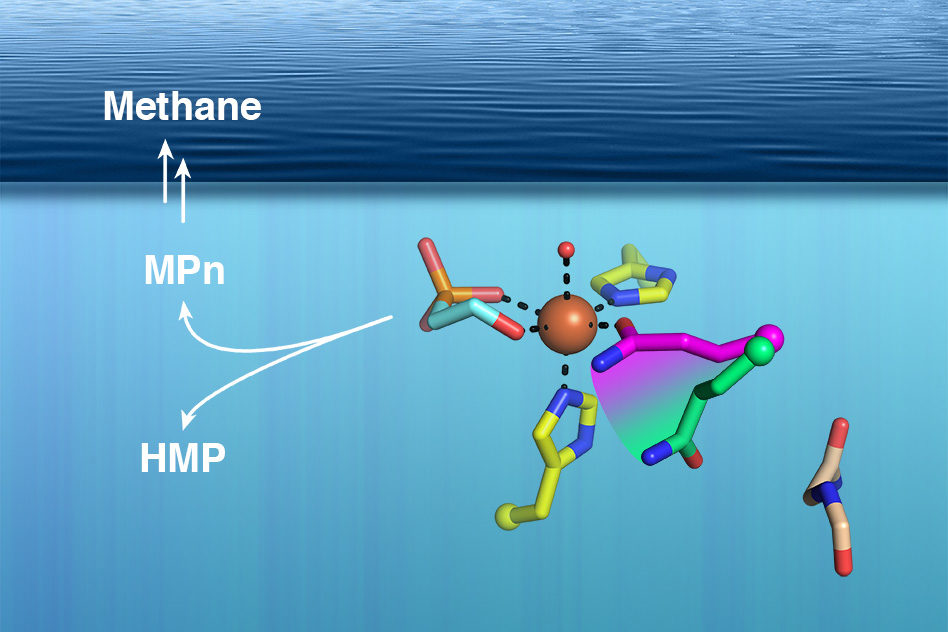
First, they identified the structure of an enzyme that can produce a compound that is known to be converted to methane. Then, they used that information to show that this enzyme exists in some of the most abundant marine microbes. They believe that this compound is likely the source of methane gas being released into the atmosphere above the ocean.
Ocean-produced methane represents around 4% of the total that is discharged into the atmosphere, and a better understanding of where this methane is coming from could help scientists better account for its role in climate change, the researchers say.
“Understanding the global carbon cycle is really important, especially when talking about climate change,” said Catherine Drennan, MIT professor of chemistry and biology and Howard Hughes Medical Institute Investigator. “Where is methane really coming from? How is it being used? Understanding nature’s flux is important information to have in all of those discussions.”
Drennan and Wilfred van der Donk, a professor of chemistry at the University of Illinois at Urbana-Champaign, are the senior authors of the paper published in the journal Science. Lead authors are David Born, a graduate student at MIT and Harvard University, and Emily Ulrich, a graduate student at the University of Illinois at Urbana-Champaign.
Many bacteria produce methane as a byproduct of their metabolism, but most of these bacteria live in oxygen-poor environments such as the deep ocean or the digestive tract of animals — not near the ocean’s surface.
Several years ago, van der Donk and University of Illinois colleague William Metcalf found a possible clue to the mystery of ocean methane: They discovered a microbial enzyme that produces a compound called methylphosphonate, which can become methane when a phosphate molecule is cleaved from it. This enzyme was found in a microbe called Nitrosopumilus maritimus, which lives near the ocean surface, but the enzyme was not readily identified in other ocean microbes as one would have expected it to be.
Van der Donk’s team knew the genetic sequence of the enzyme, known as methylphosphonate synthase (MPnS), which allowed them to search for other versions of it in the genomes of other microbes. However, every time they found a potential match, the enzyme turned out to be a related enzyme called hydroxyethylphosphonate dioxygenase (HEPD), which generates a product that is very similar to methylphosphonate but cannot be cleaved to produce methane.
Van der Donk asked Drennan, an expert in determining chemical structures of proteins, if she could try to reveal the structure of MPnS, in hopes that it would help them find more variants of the enzyme in other bacteria.
To find the structure, the MIT team employed x-ray crystallography, which they performed in a special chamber with no oxygen at the Northeastern Collaborative Access Team 24-ID-C x-ray beamline at the APS (the APS is an Office of Science user facility). They knew that the enzyme requires oxygen to catalyze the production of methylphosphonate, so by eliminating oxygen they were able to get snapshots of the enzyme as it bound to the necessary reaction partners but before it performed the reaction.
The researchers compared the crystallography data from MPnS with the related HEPD enzyme and found one small but critical difference. In the active site of both enzymes (the part of the protein that catalyzes chemical reactions), there is an amino acid called glutamine. In MPnS, this glutamine molecule binds to iron, a necessary cofactor for the production of methylphosphonate. The glutamine is fixed in an iron-binding orientation by the bulky amino acid isoleucine, which is directly below the glutamine in MPnS. However, in HEPD, the isoleucine is replaced by glycine, and the glutamine is free to rearrange so that it is no longer bound to iron.
“We were looking for differences that would lead to different products, and that was the only difference that we saw,” Born said. Furthermore, the researchers found that changing the glycine in HEPD to isoleucine was sufficient to convert the enzyme to an MPnS.
By searching databases of genetic sequences from thousands of microbes, the researchers found hundreds of enzymes with the same structural configuration seen in their original MPnS enzyme. Furthermore, all of these were found in microbes that live in the ocean, and one was found in a strain of an extremely abundant ocean microbe known as Pelagibacter ubique.
“This exciting result builds on previous, related studies showing that the metabolism of the methylphosphonate can lead to the formation of methane in the oxygenated ocean. Since methane is a potent greenhouse gas with poorly understood sources and sinks in the surface ocean, the results of this study will serve to facilitate a more comprehensive understanding of the methylphosphonate cycle in nature,” said David Karl, a professor of oceanography at the University of Hawaii, who was not involved in the research.
It is still unknown what function the MPnS enzyme and its product serve in ocean bacteria. Methylphosphonates are believed to be incorporated into fatty molecules called phosphonolipids, which are similar to the phospholipids that make up cell membranes.
“The function of these phosphonolipids is not well-established, although they’ve been known to be around for decades. That’s a really interesting question to ask,” Born said. “Now we know they’re being produced in large quantities, especially in the ocean, but we don’t actually know what they do or how they benefit the organism at all.”
Another key question is how the production of methane by these organisms is influenced by environmental conditions in the ocean, including temperature and pollution such as fertilizer runoff.
“We know that methylphosphonate cleavage occurs when microbes are starved for phosphorus, but we need to figure out what nutrients are connected to this, and how is that connected to the pH of the ocean, and how is it connected to temperature of the ocean,” Drennan said. “We need all of that information to be able to think about what we’re doing, so we can make intelligent decisions about protecting the oceans.”
See: David A. Born1,2, Emily C. Ulrich3, Kou-San Ju3,4, Spencer C. Peck3, Wilfred A. van der Donk3**, Catherine L. Drennan2*, “Structural basis for methylphosphonate biosynthesis,” Science 358, 1336 (8 December 2017). DOI: 10.1126/science.aao3435
Author affiliations: 1Harvard University, 2Massachusetts Institute of Technology, 3University of Illinois at Urbana–Champaign, 4The Ohio State University
Correspondence: *[email protected], **[email protected]
This work was supported by the National Institutes of Health (NIH, GM 077596 to W.A.V.). An NIH Molecular Biophysics Training Grant T32 GM008313 supported D.A.B. C.L.D. and W.A.V. are Howard Hughes Medical Institute Investigators. This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P41 GM103403). The Pilatus 6M detector on 24-ID-C beam line is funded by a NIH-ORIP HEI grant (S10 RR029205). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
This research used resources of the Advanced Photon Source, a U.S. DOE Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02- 06CH11357.
Argonne National Laboratory seeks solutions to pressing national problems in science and technology. The nation's first national laboratory, Argonne conducts leading-edge basic and applied scientific research in virtually every scientific discipline. Argonne researchers work closely with researchers from hundreds of companies, universities, and federal, state and municipal agencies to help them solve their specific problems, advance America's scientific leadership and prepare the nation for a better future. With employees from more than 60 nations, Argonne is managed by UChicago Argonne, LLC for the U.S. Department of Energy's Office of Science.
The U.S. Department of Energy's Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time. For more information, visit the Office of Science website.
