 Biosynthetic engineering of novel molecules to create new potential medical treatments often involves tinkering with existing natural molecules, such as antibiotics or peptides, to make them better. This can be done with standard synthetic chemical reactions in the laboratory but sometimes it is easier to find an enzyme to do the work. The search for such an enzyme has led a group of researchers down an unlikely path. By employing high-brightness x-rays from the U.S. Department of Energy’s Advanced Photon Source (APS), an Office of Science user facility, to study a strain of bacteria isolated from a coal mine fire in Appalachia, the team has identified an enzyme that catalyzes a rare reaction and applied it to making a new, improved antibiotic.
Biosynthetic engineering of novel molecules to create new potential medical treatments often involves tinkering with existing natural molecules, such as antibiotics or peptides, to make them better. This can be done with standard synthetic chemical reactions in the laboratory but sometimes it is easier to find an enzyme to do the work. The search for such an enzyme has led a group of researchers down an unlikely path. By employing high-brightness x-rays from the U.S. Department of Energy’s Advanced Photon Source (APS), an Office of Science user facility, to study a strain of bacteria isolated from a coal mine fire in Appalachia, the team has identified an enzyme that catalyzes a rare reaction and applied it to making a new, improved antibiotic.
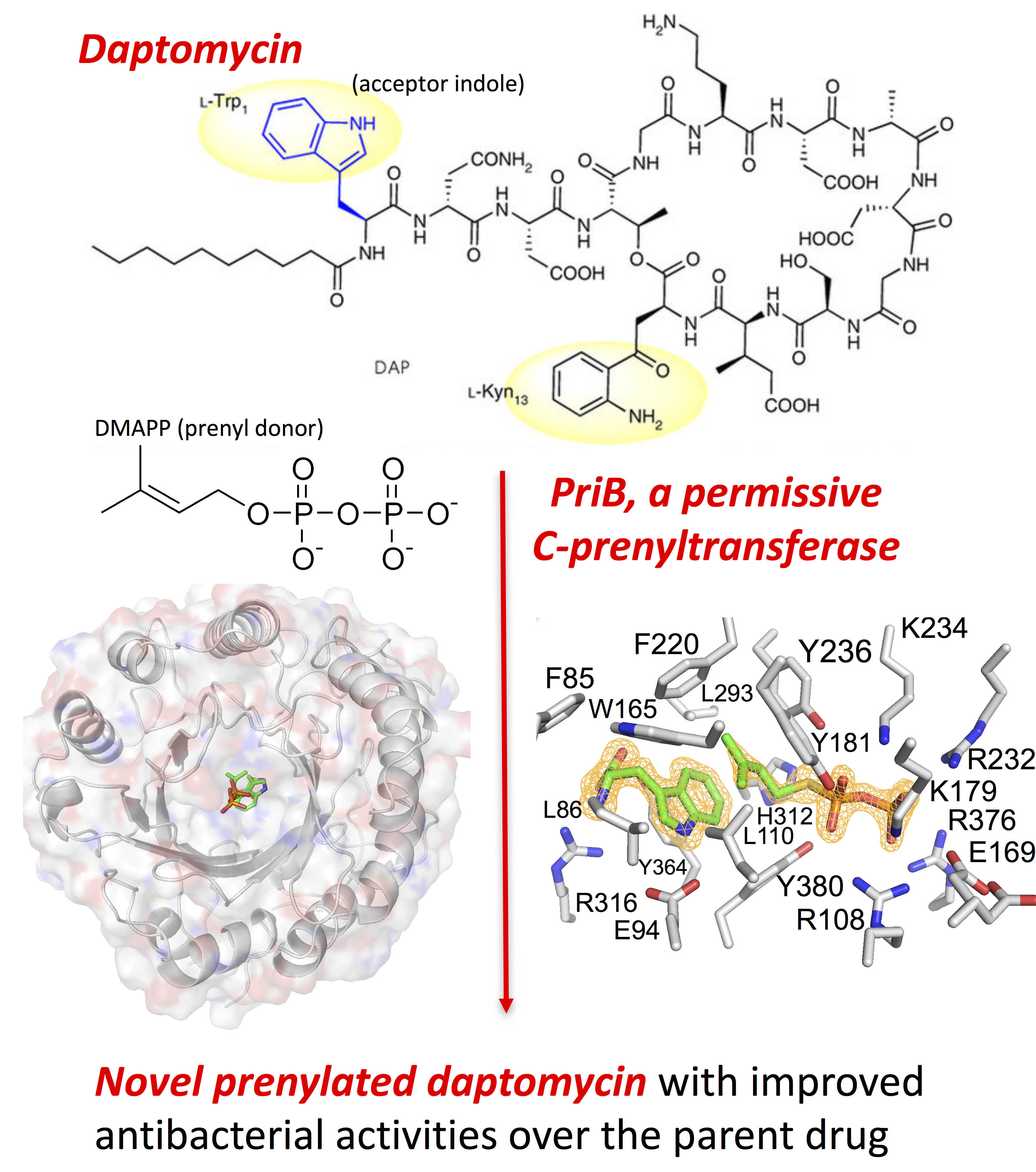
The research was based on the observation that a bacterial strain isolated from an underground coal mine fire in Kentucky was able to synthesize two distinct classes of a rare unsaturated hexuronic acid metabolite, suggesting that the bacterium expresses an enzyme that catalyzes a reaction that has presented a challenge to synthetic chemists. In order to identify the enzyme, the team from the University of Kentucky and Rice University sequenced the entire genome of the bacterial strain and performed a bioinformatics analysis on the data to identify the genes. This analysis identified a potential candidate, a single prenyl transferase gene, PriB, that clusters with other biosynthetic genes responsible for making the unusual metabolite.
In order to test whether the enzyme was indeed responsible for the reaction they were interested in, they synthesized the protein in a bacterial expression system and purified it for enzymatic testing. The enzyme assays revealed that PriB was the gene they were seeking and that it is a particularly permissive prenyl transferase that is able to act on a wider variety of substrates than other prenyl transferases in the family, with a wide range of active pH and temperature conditions, and no dependence on metals. Interestingly, the enzyme works on both D- and L-tryptophan, a first for this family of enzymes.
The ability of PriB to work on a wide range of substrates under a wide range of conditions suggested to the researchers that it could be a very useful tool for synthesizing new and interesting molecules from existing natural products. To test one of these possibilities, they tried modifying daptomycin, an antibiotic that contains the right chemical structure to be modified by PriB. It worked! PriB was able to modify daptomycin to make two new products that work 6-10 times better than daptomycin (Fig. 1).
To investigate the structural basis for the permissiveness of PriB, the team solved X-ray crystal structures of PriB with and without ligands. Analysis of data collected at the Lilly Research Laboratory Collaborative Access Team 31-ID-D x-ray beamline at the APS at Argonne National Laboratory revealed a central barrel structure around a solvent-filled core surrounded by a ring of solvent-exposed alpha helices (Fig. 1). The structure undergoes significant conformational changes upon substrate binding and researchers were able to identify differences in PriB that explain why its catalytic action is not dependent on metals the way other prenyl transferases in the family are. PriB also has a unique substrate orientation and binding pocket dynamics that explain its substrate permissiveness compared to other members of the family.
The discovery and characterization of PriB opens doors to new possibilities for biosynthetic tailoring of natural products to make new, more effective products and treatments. The modification of daptomycin in this study is sufficient proof that new tools can be found in the most unlikely locations, even underground in a coal mine fire. — Sandy Field
See: Sherif I. Elshahawi1, Hongnan Cao2, Khaled A. Shaaban1, Larissa V. Ponomareva1, Thangaiah Subramanian1, Mark L. Farman1, H. Peter Spielmann1, George N. Phillips, Jr.,2*, Jon S. Thorson1,** and Shanteri Singh1***‡, “Structure and specificity of a permissive bacterial C-prenyltransferase,” Nat. Chem. Biol. 13, 366 (April 2017). DOI: 10.1038/nchembio.2285
Author affiliations: 1University of Kentucky, 2Rice University ‡Present address: University of Oklahoma
Correspondence: *[email protected], **[email protected], ***[email protected]
This work was supported by NIH grants R37 AI52188 and R01 CA203257 (J.S.T.), U01 GM098248 (G.N.P.) and NCATS (UL1TR001998). Use of the Lilly Research Laboratories Collaborative Access Team beamline at Sector 31 of the Advanced Photon Source was provided by Eli Lilly and Company. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science user facility operated by Argonne National Laboratory (DE-AC02-06CH11357).
Argonne National Laboratory seeks solutions to pressing national problems in science and technology. The nation's first national laboratory, Argonne conducts leading-edge basic and applied scientific research in virtually every scientific discipline. Argonne researchers work closely with researchers from hundreds of companies, universities, and federal, state and municipal agencies to help them solve their specific problems, advance America's scientific leadership and prepare the nation for a better future. With employees from more than 60 nations, Argonne is managed by UChicago Argonne, LLC for the U.S. Department of Energy's Office of Science.
The U.S. Department of Energy's Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time. For more information, visit the Office of Science website.
