The original Penn State press release by Gail McCormick can be read here.
Images of a protein involved in creating a potent antibiotic reveal the unusual first steps of the antibiotic’s synthesis. The improved understanding of the chemistry behind this process is detailed in a new study led by Penn State chemists based upon research at two U.S. Department of Energy x-ray light sources, including the Advanced Photon Source, and published in the journal Nature Chemical Biology. These results could allow researchers to adapt this and similar compounds for use in human medicine.
“The antibiotic thiostrepton is very potent against Gram-positive pathogens and can even target certain breast cancer cells in culture,” said Squire Booker, a biochemist at Penn State and investigator with the Howard Hughes Medical Institute. “While it has been used topically in veterinary medicine, so far it has been ineffective in humans because it is poorly absorbed. We studied the first steps in thiostrepton’s biosynthesis in hopes of eventually being able to hijack certain processes and make analogs of the molecule that might have better medicinal properties. Importantly, this reaction is found in the biosynthesis of numerous other antibiotics, and so the work has the potential to be far reaching.”
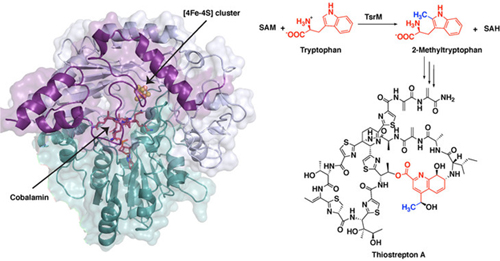
The first step in thiostrepton’s synthesis involves a process called methylation. A molecular tag called a methyl group, which is important in many biological processes, is added to a molecule of tryptophan, the reaction’s substrate. One of the major systems for methylating compounds that are not particularly reactive, like tryptophan, involves a class of enzymes called radical SAM proteins.
“Radical SAM proteins usually use an iron-sulfur cluster to cleave a molecule called S-adenosyl-L-methionine (SAM), producing a “free radical” or an unpaired electron that helps move the reaction forward,” said Hayley Knox, a graduate student in chemistry at Penn State and first author of the paper. “The one exception that we know about so far is the protein involved in the biosynthesis of thiostrepton, called TsrM. We wanted to understand why TsrM doesn’t do radical chemistry, so we used an imaging technique called x-ray crystallography to investigate its structure at several stages throughout its reaction.” The x-ray crystallography experiments were carried out at the General Medical Sciences and Cancer Institutes (GM/CA-XSD) and Life Sciences Collaborative Access Team (LS-CAT) x-ray beamlines at the Advanced Photon Source at Argonne National Laboratory, and at the Berkeley Center for Structural Biology beamlines at the Advanced Light Source at Lawrence Berkeley National Laboratory. The Advanced Photon Source and the Advanced Light Source are U.S. Department of Energy user facilities.
In all radical SAM proteins characterized to date, SAM binds directly to the iron-sulfur cluster, which helps to fragment the molecule to produce the free radical. However, the researchers found that the site where SAM would typically bind is blocked in TsrM.
“This is completely different from any other radical SAM protein,” said Booker. “Instead, the portion of SAM that binds to the cluster associates with the tryptophan substrate and plays a key role in the reaction, in what is called substrate-assisted catalysis.”
In solving the structure, the researchers from the Massachusetts Institute of Technology and the Albert Einstein College of Medicine, in addition to Pennsylvania State University, were able to infer the chemical steps during the first part of thiostrepton’s biosynthesis, when tryptophan is methylated. In short, the methyl group from SAM transfers to a part of TsrM called cobalamin. Then, with the help of an additional SAM molecule, the methyl group transfers to tryptophan, regenerating free cobalamin and producing the methylated substrate, which is required for the next steps in synthesizing the antibiotic.
“Cobalamin is the strongest nucleophile in nature, which means it is highly reactive,” said Knox. “But the substrate tryptophan is weakly nucleophilic, so a big question is how cobalamin could ever be displaced. We found that an arginine residue sits under the cobalamin and destabilizes the methyl-cobalamin, allowing tryptophan to displace cobalamin and become methylated.”
Next the researchers plan to study other cobalamin-dependent radical SAM proteins to see if they operate in similar ways. Ultimately, they hope to find or create analogs of thiostrepton that can be used in human medicine.
“TsrM is clearly unique in terms of known cobalamin-dependent radical SAM proteins and radical SAM proteins in general,” said Booker. “But there are hundreds of thousands of unique sequences of radical SAM enzymes, and we still don’t know what most of them do. As we continue to study these proteins, we may be in store for many more surprises.”
See: Hayley L. Knox1, Percival Yang-Ting Chen2‡, Anthony J. Blaszczyk1‡‡, Arnab Mukherjee1, Tyler L. Grove3, Erica L. Schwalm1‡‡‡, Bo Wang1, Catherine L. Drennan2*, and Squire J. Booker1**, “Structural basis for non-radical catalysis by TsrM, a radical SAM methylase,” Nat. Chem. Biol., published on line 18 January 2021. DOI: 10.1038/s41589-020-00717-y
Author affiliations: 1Pennsylvania State University, 2Massachusetts Institute of Technology, 3Albert Einstein College of Medicine Present addresses: ‡University of California, San Diego, ‡‡Catalent Pharma Solutions, ‡‡‡Merck & Co., Inc.
Correspondence: *[email protected], **[email protected]
This work was supported by the National Institutes of Health (NIH) GM-122595 to S.J.B. and GM-126982 to C.L.D., and the Eberly Family Distinguished Chair in Science (S.J.B.). S.J.B. and C.L.D. are investigators of the Howard Hughes Medical Institute. Use of GM/CA-XSD has been funded in whole or in part with federal funds from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006). The Eiger 16M detector at GM/CA-XSD was funded by NIH grant S10 OD012289. Use of the LS-CAT at Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (grant 085P1000817). This research also used the resources of the Berkeley Center for Structural Biology supported in part by the Howard Hughes Medical Institute. The Advanced Light Source is a U.S. Department of Energy (DOE) Office of Science User Facility under contract no. DE-AC02-05CH11231. The ALS-ENABLE beamlines are supported in part by the NIH, National Institute of General Medical Sciences grant P30 GM124169. This research used resources of the Advanced Photon Source, a U.S. DOE Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under contract no. DE-AC02-06CH11357.
The Pennsylvania State University © 2021
The U.S. Department of Energy's APS is one of the world’s most productive x-ray light source facilities. Each year, the APS provides high-brightness x-ray beams to a diverse community of more than 5,000 researchers in materials science, chemistry, condensed matter physics, the life and environmental sciences, and applied research. Researchers using the APS produce over 2,000 publications each year detailing impactful discoveries, and solve more vital biological protein structures than users of any other x-ray light source research facility. APS x-rays are ideally suited for explorations of materials and biological structures; elemental distribution; chemical, magnetic, electronic states; and a wide range of technologically important engineering systems from batteries to fuel injector sprays, all of which are the foundations of our nation’s economic, technological, and physical well-being.
Argonne National Laboratory seeks solutions to pressing national problems in science and technology. The nation's first national laboratory, Argonne conducts leading-edge basic and applied scientific research in virtually every scientific discipline. Argonne researchers work closely with researchers from hundreds of companies, universities, and federal, state and municipal agencies to help them solve their specific problems, advance America's scientific leadership and prepare the nation for a better future. With employees from more than 60 nations, Argonne is managed by UChicago Argonne, LLC, for the U.S. DOE Office of Science.
The U.S. Department of Energy's Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time. For more information, visit the Office of Science website.
