The original Salk News article can be read here
In structural biology, some molecules are so unusual they can only be captured with a unique set of tools. That’s precisely how a multi-institutional research team led by Salk scientists defined how antibodies can recognize a compound called phosphohistidine—a highly unstable molecule that has been found to play a central role in some forms of cancer, such as liver and breast cancer and neuroblastoma. These insights not only set up the researchers for more advanced studies on phosphohistidine and its potential role in cancer, but will also enable scientists to manipulate the shape and atomic makeup of the antibodies’ binding sites in order to design ever more efficient antibodies in the future. The study, using data obtained at three U.S. Department of Energy x-ray light sources including the Advanced Photon Source, was published in the Proceedings of the National Academy of Sciences of the United States of America.
“We are excited that these new antibody structures reveal novel principles of antigen binding. Now we can redesign these antibodies and engineer their properties to make them more efficient,” said Tony Hunter, Renato Dulbecco Chair and American Cancer Society Professor at Salk and the paper’s senior author. “This work may also provide other scientists with phosphohistidine antibodies that better suit their research purposes.”
Amino acids are joined together in precise sequences to form proteins, and several of them can undergo chemical transformations that can change the activity of the protein for better or worse. One such transformation is a process called phosphorylation, when a compound called phosphate is added to an amino acid, changing its shape and charge. Previously, Hunter showed that phosphorylation on the amino acid tyrosine can drive cancer progression, a discovery that led to numerous anticancer drugs. More recently, Hunter turned his attention to phosphorylation of the amino acid histidine (which creates phosphohistidine), suspecting that the process might also play a role in human disease.
Hunter developed a suite of antibodies able to bind to phosphohistidine in proteins, and used chemically stabilized phosphohistidine analogues to develop a series of monoclonal antibodies that could recognize these forms. The next step was to understand exactly how the antibodies are able to bind to phosphohistidine. This led Hunter to collaborate with Ian Wilson, the Hansen Professor of Structural Biology at the Scripps Research Institute and a world-renowned expert in using protein crystallography to define antibody structures, to study the structures of the phosphohistidine antibodies.
“My long-term colleague Tony and I have been collaborating on this project for the past seven years,” says Wilson. “We have obtained new insights into how antibodies can evolve to recognize phosphates linked to proteins, which is very satisfying.”
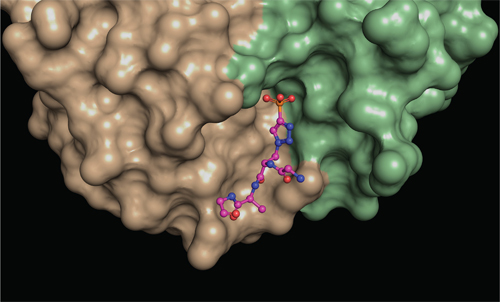
To find out how phosphohistidine is recognized, they needed to image their antibodies in the act of binding the phosphohistidine, and so formed crystals between each antibody bound to a phosphohistidine peptide.
“To understand the molecular interactions between the antibodies and phosphohistidine, we needed to look at them in great detail,” says first author Rajasree Kalagiri, a Salk postdoctoral researcher and expert in x-ray crystallography. “Once we got the antibodies to form crystals, we bombarded those crystals with x-rays to obtain a diffraction pattern. We then applied methods that transform the diffraction pattern into a three-dimensional electron density map, which was then used to discern the atomic structure of the antibodies.”
The x-ray diffraction patterns were determined at the National Institute of General Medical Sciences and National Cancer Institute Structural Biology Facility (GM/CA-XSD) 23-ID-D beamline at the Argonne National Laboratory Advanced Photon Source, the Stanford Synchrotron Radiation Lightsource beamline 12-2 at the SLAC National Accelerator Laboratory, and the Advanced Light Source beamline 5.0.3 at Lawrence Berkeley National Laboratory. All three are DOE Office of Science user facilities.
The two types of antibody crystal structures solved by the team revealed exactly how different amino acids are arranged around the phosphohistidine to bind it tightly. Their five structures more than double the number of phospho-specific antibody structures previously reported and provide insights into how antibodies recognize both the phosphate and the linked histidine. They also reveal at a structural level how the two types of antibody recognize different forms of phosphohistidine, and this will allow the scientists to engineer improved antibodies in the future.
See: Rajasree Kalagiri1, Robyn L. Stanfield2, Jill Meisenhelder1, James J. La Clair1,3, Stephen R. Fuhs1‡, Ian A. Wilson2, and Tony Hunter1,2, “Structural basis for differential recognition of phosphohistidine-containing peptides by 1-pHis and 3-pHis monoclonal antibodies,” Proc. Natl. Acad. Sci. U.S.A., 118(6), e2010644118 (2021). DOI: 10.1073/pnas.2010644118
Author affiliations: 1Salk Institute for Biological Studies, 2The Scripps Research Institute, 3University of California San Diego ‡Present address: Genomics Institute of the Novartis Research Foundation
Correspondence: * [email protected]
Use of the Stanford Synchrotron Radiation Light source, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy (DOE) Office of Science-Basic Energy Sciences, under Contract DE-AC02-76SF00515. The Advanced Light Source is a DOE Office of Science User Facility under Contract DE-AC02-05CH11231. The Pilatus detector on 5.0.1. was funded under National Institutes of Health (NIH) Grant S10OD021832. GM/CA-XSD has been funded by the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006, P30GM138396). Funding for reagents used in this project, and salary support, was provided by NIH 5 R01 CA082683, NIH 5 R01 CA194584, and NIH 1 R35 CA242443 (to T.H.); Leona M. and Harry B. Helmsley Charitable Trust Grant 2012-PG-MED002; and the Skaggs Institute for Chemical Biology at The Scripps Research Institute. The Salk Peptide Synthesis and Proteomics Cores are supported by P30 CA014195. T.H. is a Frank and Else Schilling American Cancer Society Professor and holds the Renato Dulbecco Chair in Cancer Research. I.A.W. is the Hansen Professor of Structural Biology. This research used resources of the Advanced Photon Source, a U.S. DOE Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357.
Copyright 2021 Salk Institute for Biological Studies
The U.S. Department of Energy's APS is one of the world’s most productive x-ray light source facilities. Each year, the APS provides high-brightness x-ray beams to a diverse community of more than 5,000 researchers in materials science, chemistry, condensed matter physics, the life and environmental sciences, and applied research. Researchers using the APS produce over 2,000 publications each year detailing impactful discoveries, and solve more vital biological protein structures than users of any other x-ray light source research facility. APS x-rays are ideally suited for explorations of materials and biological structures; elemental distribution; chemical, magnetic, electronic states; and a wide range of technologically important engineering systems from batteries to fuel injector sprays, all of which are the foundations of our nation’s economic, technological, and physical well-being.
Argonne National Laboratory seeks solutions to pressing national problems in science and technology. The nation's first national laboratory, Argonne conducts leading-edge basic and applied scientific research in virtually every scientific discipline. Argonne researchers work closely with researchers from hundreds of companies, universities, and federal, state and municipal agencies to help them solve their specific problems, advance America's scientific leadership and prepare the nation for a better future. With employees from more than 60 nations, Argonne is managed by UChicago Argonne, LLC, for the U.S. DOE Office of Science.
The U.S. Department of Energy's Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time. For more information, visit the Office of Science website.
