Antibiotic resistance is a global problem that many scientists are racing to solve. Bacterial infections that were once easily controlled by penicillin or tetracycline are now impossible to treat except with our most recently developed antimicrobials. This is due to the fact that bacteria can develop, and spread/disperse, mutations that allow them to evade the effects of antibiotics. One type of bacteria that has been very successful at this is Mycobacterium tuberculosis, a deadly bacterium responsible for the deaths of ~1.5 million people in 2018. Multi-drug resistant and extensively drug-resistant strains of M. tuberculosis have become a major global problem, increasing the need for new antibiotics. This has driven researchers to seek new bacterial targets for structure-based drug design. In recent work conducted at the U.S. Department of Energy’s Advanced Photon Source (APS), researchers gained important insights into understanding the structure and mechanism of a critical enzyme involved in protein synthesis in M. tuberculosis. Their work, published in Nucleic Acids Research, will form the basis of structure-based drug design efforts aimed at exploiting the unique features of this enzyme, a method that has been successful in developing pharmaceuticals against methicillin-resistant Staphylococcus aureus (MRSA) and some fungal infections.
The research focuses on learning more about an essential enzyme involved in making proteins from a messenger RNA (mRNA) transcript. This is a basic function of all cells, in both bacteria and multi-cellular organisms, and involves a coordinated set of specific enzymes and special RNA molecules called transfer RNAs (tRNAs). The tRNAs show “L” shape and they recognize a 3-base sequence in the mRNA called a codon and translate it into the correct amino acid code using a matching anticodon sequence. A set of 20 critical enzymes in this process are the aminoacyl tRNA synthetases (aaRS). Each aaRS loads the specific amino acid onto correct tRNA molecule in a two-step reaction. One important consideration is that, although aaRSs function in a very evolutionarily conserved process that all organisms carry out, these enzymes have diverged structurally enough make the ones in bacteria distinct from our own so we can target them without disrupting protein synthesis in our own cells.
The work was done as a collaboration between researchers at Argonne National Laboratory, The University of Chicago, Auburn University, and the University of Dundee (Scotland) and involved structural analysis of the M. tuberculosis aaRS, called MtFRS, that transfers the amino acid phenylalanine to its specific tRNA, tRNAPhe. In order to join the correct amino acid to its appropriate tRNA, the enzyme must recognize specific features of tRNAPhe that distinguish it from the tRNAs for the other 19 amino acids and then bring the phenylalanine and tRNAPhe into proximity to catalyze the ligation reaction.
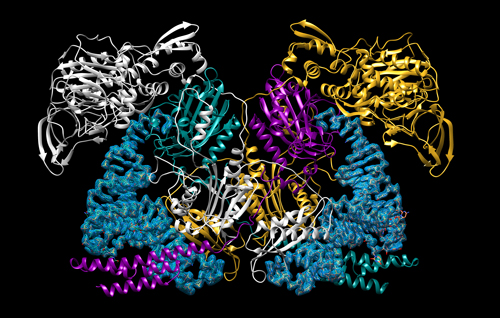
Using the Structural Biology Center (SBC-XSD) x-ray beamline 19-IDS at the APS, the team solved four structures of large complexes that capture the two steps of the MtFRS enzyme’s catalytic action and tRNA recognition (Fig. 1). The initial binding of the amino acid was demonstrated by two crystal structures of MtFRS bound to M. tuberculosis tRNAPhe (MttRNAPhe) with phenylalanine and one with a non-hydrolyzable phenylalanine analog. The aminoacylation “ready” state, in which the tRNA is set to accept loading of the amino acid, was demonstrated by crystallization of MtFRS bound to MttRNAPhe with an inhibitor, the non-hydrolyzable analog of amino acid.
The structures confirmed previous work showing that MtFRS adopts a large and complex heterotetrameric structure and it binds two molecules of MttRNAPhe. In this work, the team discovered that initial recognition of the tRNAPhe is performed by one half of the heterotetramer while aminoacylation is performed by the other half. Selective recognition of the tRNA is provided by enzyme interactions with the anticodon loop and stem regions of the tRNAPhe but, surprisingly, not the unique anticodon sequence itself. Instead, MtFRS appears to recognize only the wobble base, the least important aspect of recognition between the tRNAPhe anticodon and mRNA codon sequence. These interactions are maintained in the ready state and this work reported, for the first time, the process of stabilization and ordering of the active site that brings the correct catalytic groups into the appropriate geometry for the aminoacylation reaction, confirming the proposed catalytic mechanism of MtFRS. Comparison of these structures to those of human aaRSs and those of other bacteria allowed the team to identify some unique features of MtFRS that provide a good starting point for the design of small molecule inhibitors to fight tuberculosis infection. ― Sandy Field
See: Karolina Michalska1,2, Robert Jedrzejczak1,2, Jacek Wower3, Changsoo Chang1,2, Beatriz Baragaña4, Ian H. Gilbert4, Barbara Forte4, and Andrzej Joachimiak1,2*, “Mycobacterium tuberculosis Phe-tRNA synthetase: structural insights into tRNA recognition and aminoacylation,” Nucleic Acids Res. 1, 5351 (21 May, 2021). DOI: 10.1093/nar/gkab272
Author affiliations: 1The University of Chicago, 2Argonne National Laboratory, 3Auburn University, 4University of Dundee
Correspondence: * [email protected]
The authors thank the members of the SBC-XSD at Argonne National Laboratory for their help with setting beamline and data collection at beamline 19-ID. Funding: National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services [HHSN272201200026C, HHSN272201700060C, in part]; U.S. Department of Energy (DOE) Office of Science and operated for the DOE Office of Science by Argonne National Laboratory [DE-AC02-06CH11357]; J.W. was supported by the Hatch program of the National Institute of Food and Agriculture, U.S. Department of Agriculture. Funding for open access charge: National Institutes of Health. SBC-XSD is operated by UChicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357.
The U.S. Department of Energy's APS is one of the world’s most productive x-ray light source facilities. Each year, the APS provides high-brightness x-ray beams to a diverse community of more than 5,000 researchers in materials science, chemistry, condensed matter physics, the life and environmental sciences, and applied research. Researchers using the APS produce over 2,000 publications each year detailing impactful discoveries, and solve more vital biological protein structures than users of any other x-ray light source research facility. APS x-rays are ideally suited for explorations of materials and biological structures; elemental distribution; chemical, magnetic, electronic states; and a wide range of technologically important engineering systems from batteries to fuel injector sprays, all of which are the foundations of our nation’s economic, technological, and physical well-being.
Argonne National Laboratory seeks solutions to pressing national problems in science and technology. The nation's first national laboratory, Argonne conducts leading-edge basic and applied scientific research in virtually every scientific discipline. Argonne researchers work closely with researchers from hundreds of companies, universities, and federal, state and municipal agencies to help them solve their specific problems, advance America's scientific leadership and prepare the nation for a better future. With employees from more than 60 nations, Argonne is managed by UChicago Argonne, LLC, for the U.S. DOE Office of Science.
The U.S. Department of Energy's Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time. For more information, visit the Office of Science website.
