When SARS-CoV-2, the virus responsible for the disease COVID-19, infects a person’s cells, it provides the instructions to make the proteins responsible for viral replication and release. An essential step in this process is the cleaving of a long viral polyprotein into smaller functional units, remarkably carried out in part by an enzyme, Mpro, itself contained within the polyprotein. Researchers using the U.S. Department of Energy’s (DOE’s) Advanced Photon Source (APS) have determined crystal structures that capture the moment Mpro begins this process. Aside from benefits to basic research, the structures will help guide the researchers in developing small-molecule drugs designed to stop the spread of COVID-19 by inhibiting Mpro’s ability to set viral replication in motion. These results were published in the journal Nature Communications.
Coronaviruses became a major public health concern in 2002 when severe acute respiratory syndrome coronavirus (SARS-CoV) jumped from animals to humans, in a process known as “spillover”, where it caused SARS, a severe respiratory illness. This was followed in 2012 by the emergence of Middle East respiratory syndrome, caused by MERS-CoV, and COVID-19 in 2019, caused by SARS-CoV-2. Far more transmissible than either SARS-CoV or MERS-CoV, SARS-CoV-2 has caused nearly 2.5 million deaths worldwide to date, making it a global priority.
SARS-CoV-2 is a positive-sense, single-stranded RNA virus, meaning its genetic material can be directly translated into proteins in a host’s cell. One of these, replicase polyprotein pp1a/ab, is a single polypeptide that is “chopped up” into 16 proteins, which are involved in viral replication within the host. Remarkably, the enzymes responsible for this—PLpro and Mpro—are themselves contained within the large polyprotein and must cut themselves from the polypeptide at their own N- and C- termini, prior to releasing the other proteins. If this can be prevented, replication of the virus will not take place.
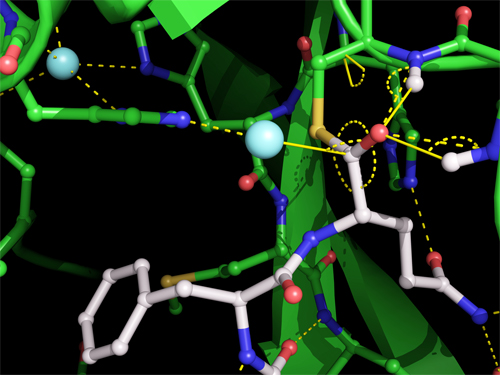
To that end, researchers from the University of British Columbia and Simon Fraser University (both Canada) honed in on Mpro’s method of excising itself from the polypeptide, especially the moment Mpro binds and cuts its C-terminus. This transitory event creates a temporary structure called the acyl-enzyme intermediate, pictured in Fig. 1.
To achieve the crystal structures, the researchers generated the protein in E. coli at the University of British Columbia, where they purified it, prepared the crystals, and screened them with an in-house Rigaku x-ray generator. After cryo-protection in liquid nitrogen, they sent the crystals to the APS. Using macromolecular x-ray crystallography at the National Institute of General Medical Sciences and National Cancer Institute (GM/CA-XSD) structural biology facility x-ray beamline 23-ID-B at the APS (a DOE Office of Science user facility at Argonne National Laboratory) the researchers determined three crystal structures: Mpro in its unbound state, at 1.8-Å resolution; the acyl-enzyme intermediate, at 1.95 Å; and the substrate bound to a mutant Mpro engineered to prevent the cut, at 2.0-Å resolution.
The crystal structure of the acyl-enzyme intermediate is the first one scientists have trapped for a coronavirus protease and contributes valuable information for the design of anti-viral drugs. It shows that Mpro forms a covalent link between its catalytic cysteine side chain and the carbonyl carbon of the substrate, with a water molecule positioned to make the excising cut. The carbonyl oxygen is stabilized by hydrogen bonds with main chain nitrogens, forming an oxyanion hole, commonly found in enzyme intermediates. By comparing the crystal of the acyl-enzyme intermediate with the other two, researchers are able to ascertain changes in the width and shape in Mpro’s substrate binding groove as it shifts to accommodate its substrate; the precise mode of binding; and the potential angle of action of the deacylating water molecule.
The APS offered the researchers three particular assets: (1) the high intensity and small size of the x-ray beam allowed them to work with small crystals; (2) the high intensity further enabled diffraction to high resolution, which permitted precise location of the protein atoms and water molecules that are critical for mechanistic understanding; and (3) the tunable x-ray wavelength enabled them to investigate possible metal binding sites.
The speed of data collection at the APS was also a critical factor in their work. The acyl-enzyme intermediate was identified during data collection on hundreds of crystals as part of the broader project aimed at identifying small molecules that might be candidates for anti-viral development. The x-ray beam’s short exposure time and robotically controlled loading allowed high-throughput data collection.
The crystal structures determined using data collected at the APS are so precise in their three-dimensional depiction of the acyl-enzyme intermediate that the researchers are well prepared in their quest to design a compound that can inhibit Mpro’s action. — Judy Myers
See: Jaeyong Lee1,2, Liam J. Worrall1, Marija Vuckovic1, Federico I. Rosell1, Francesco Gentile1, Anh-Tien Ton1, Nathanael A. Caveney1, Fuqiang Ban1, Artem Cherkasov1, Mark Paetzel2*, and Natalie C. J. Strynadka1**, “Crystallographic structure of wild-type SARS-CoV-2 main protease acyl-enzyme intermediate with physiological C-terminal autoprocessing site,” Nat. Commun. 11, 5877 (2020). DOI: 10.1038/s41467-020-19662-4
Author affiliations: 1The University of British Columbia, 2Simon Fraser University
Correspondence: * [email protected], ** [email protected]
GM/CA-XSD has been funded in whole or in part with Federal funds from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006); and the Eiger 16M detector by NIH grant S10 OD012289. This work was funded by operating grants from the Canadian Institutes of Health Research to N.C.J.S., M.P., and A.C. N.C.J.S. is a Tier I Canada Research Chair in Antibiotic Discovery. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. Extraordinary facility operations were supported in part by the DOE Office of Science through the National Virtual Biotechnology Laboratory, a consortium of DOE national laboratories focused on the response to COVID-19, with funding provided by the Coronavirus CARES Act.
The U.S. Department of Energy's APS is one of the world’s most productive x-ray light source facilities. Each year, the APS provides high-brightness x-ray beams to a diverse community of more than 5,000 researchers in materials science, chemistry, condensed matter physics, the life and environmental sciences, and applied research. Researchers using the APS produce over 2,000 publications each year detailing impactful discoveries, and solve more vital biological protein structures than users of any other x-ray light source research facility. APS x-rays are ideally suited for explorations of materials and biological structures; elemental distribution; chemical, magnetic, electronic states; and a wide range of technologically important engineering systems from batteries to fuel injector sprays, all of which are the foundations of our nation’s economic, technological, and physical well-being.
Argonne National Laboratory seeks solutions to pressing national problems in science and technology. The nation's first national laboratory, Argonne conducts leading-edge basic and applied scientific research in virtually every scientific discipline. Argonne researchers work closely with researchers from hundreds of companies, universities, and federal, state and municipal agencies to help them solve their specific problems, advance America's scientific leadership and prepare the nation for a better future. With employees from more than 60 nations, Argonne is managed by UChicago Argonne, LLC, for the U.S. DOE Office of Science.
The U.S. Department of Energy's Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time. For more information, visit the Office of Science website.
