Treatment for breast cancer has improved greatly in the past two decades, in part because of the recognition that about 70% of breast tumors express cellular hormone receptors called estrogen receptors (ERs) that can be targeted by a number of effective endocrine therapies. Unfortunately, after about 5 years, approximately 50% of ER-positive breast tumors will develop resistance to these treatments, leading to disease progression and metastasis. In about 25% of these cases, this resistance is due to the emergence of mutations in the ER gene (ESR1) that change the ligand binding domain (LBD) of the receptor so that it is activated even in the absence of the hormone. This has led researchers to dig deeper to try to understand the complex interplay between the structural details of the receptors and the inhibitors in order to fine tune breast cancer therapies to overcome this resistance. Recent findings based on data from studies at the U.S. Department of Energy’s Advanced Photon Source (APS) may lead to the design of new second-line breast cancer treatments or to strategies for fine-tuning the sequencing of treatments to optimize the characteristics of different drugs to avoid resistance mechanisms. These results were published in the journal eLife.
Endocrine therapies for breast cancer include those that block production of the hormones that drive ER transcriptional activity (aromatase inhibitors), those that block the receptors directly (selective ER modulators [SERMs]), and those that facilitate their cellular degradation to block their activation in cells (selective ER degrader/down regulators [SERDs]). However, although SERDs have demonstrated superior potency against ESR1-mutated breast cancer cells in culture compared to other endocrine therapies, recent data have raised questions regarding the mechanisms underlying the efficacy of these treatments against breast tumors with ESR1 mutations. In order to answer these questions, researchers from the University of Chicago, Loyola University, and the University of Montreal recently performed a study to understand more about the structures of the inhibitors used in endocrine therapy and how they modulate receptor activation and degradation.
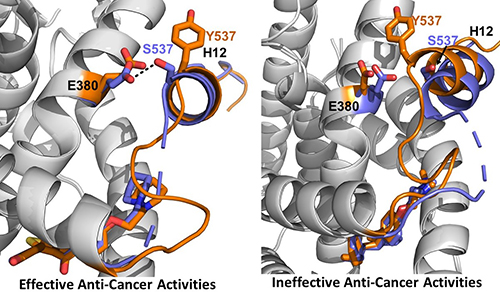
The research was conducted using a combination of cellular assays to assess ER expression and degradation, reporter gene assays to assess transcriptional activation of the receptor, and x-ray crystallography. The work focused on two mutations in the ER ligand binding domain near helix 12 (H12) that cause about 50% of ESR1-based resistance cases: Y537S and D538G (Fig. 1). The team evaluated characteristics of existing SERMs and SERDS, as well as chemically modified versions of a SERM called lasofoxifene (laso), to understand the structural features that determine how each molecule leads to inhibition, stabilization, or degradation of the ERs in breast cancer cells.
Results from the cellular degradation and transcriptional activation assays provide important insights into how both ESR1 mutations contribute to breast cancer pathology by affecting the lifetime of the ER in cells and the way the receptor responds to different SERDs and SERMs. The data suggest that the Y537S mutation activates the ER, making it resistant to SERM treatment, but also increases the number of ERs in the cell, potentially promoting tumor progression. The D538G mutation appears to dampen the effects of both SERMs and SERDs on ER degradation and to provide low-level activation of the ER in the absence of hormone. Interestingly, although the parent compound for laso did not appear to affect the degradation/accumulation of ERs in cells, two modified versions, LA-Deg and LA-Stab, enhanced degradation and stabilization, respectively, of the receptor, providing information about the structural features involved in this aspect of receptor regulation.
Using high-brightness x-ray beams from the APS at the Structural Biology Center (SBC-XSD) 19-ID beamline at the APS the team also solved x-ray crystal structures for the LBD of the Y537S ER in complex with the laso derivatives, SERDs, and SERMs and compared their results to known structures of the unmutated ER. Analysis and comparison of the data for the different structures showed that the inhibitors that favor packing of the H12 helix of the receptor into the AF-2 cleft of the protein have greater activity in terms of inhibiting the transcriptional activation of the ER (Fig. 1).
Taken together, these data provide information about the structural characteristics of endocrine inhibitors and the estrogen receptor that are important for breast cancer pathology, both in terms of activation of the receptor and in modulating ER degradation/accumulation. This information will be key to the development of new competitive anti-estrogen therapies to overcome endocrine therapy resistance in breast cancer and improve patient survival. ― Sandy Field
See: David J. Hosfield1, Sandra Weber2, Nan-Sheng Li1, Madline Suavage2, Carstyn F. Joiner3, Govinda R. Hancock3, Emily A. Sullivan3, Estelle Ndukwe1, Ross Han1, Sydney Cush1, Muriel Lainé1, Sylvie C. Mader2, Geoffrey L. Greene1, Sean W. Fanning3*, “Stereospecific lasofoxifene derivatives reveal the interplay between estrogen receptor alpha stability and antagonistic activity in ESR1 mutant breast cancer cells,” eLife 11, e72512 (2022). DOI: 10.7554/eLife.72512
Author affiliations: 1The University of Chicago, 2Université de Montréal, 3Loyola University Chicago
Correspondence: * [email protected]
Funding from Susan G. Komen Foundation CCR19608597 (SWF), Ludwig Fund for Metastasis Research (GLG), and Canadian Institutes of Health Research (SM). The Structural Biology Center (SBC-XSD) is supported by the U.S. Department of Energy (DOE) Office of Biological and Environmental Research contract DE-AC02- 06CH11257. This research used resources of the Advanced Photon Source, a U.S. DOE Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
The U.S. Department of Energy's APS at Argonne National Laboratory is one of the world’s most productive x-ray light source facilities. Each year, the APS provides high-brightness x-ray beams to a diverse community of more than 5,000 researchers in materials science, chemistry, condensed matter physics, the life and environmental sciences, and applied research. Researchers using the APS produce over 2,000 publications each year detailing impactful discoveries, and solve more vital biological protein structures than users of any other x-ray light source research facility. APS x-rays are ideally suited for explorations of materials and biological structures; elemental distribution; chemical, magnetic, electronic states; and a wide range of technologically important engineering systems from batteries to fuel injector sprays, all of which are the foundations of our nation’s economic, technological, and physical well-being.
Argonne National Laboratory seeks solutions to pressing national problems in science and technology. The nation's first national laboratory, Argonne conducts leading-edge basic and applied scientific research in virtually every scientific discipline. Argonne researchers work closely with researchers from hundreds of companies, universities, and federal, state and municipal agencies to help them solve their specific problems, advance America's scientific leadership and prepare the nation for a better future. With employees from more than 60 nations, Argonne is managed by UChicago Argonne, LLC, for the U.S. DOE Office of Science.
The U.S. Department of Energy's Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time. For more information, visit the Office of Science website.
