|
|
| Welcome to 12-ID-B |
|
12-ID-B belongs to the Chemical and Materials Science group (CMS) of X-ray Science Division at Advanced Photon Source. Beamline 12-ID-B is a dedicated X-ray scattering beamline at APS. It routinely runs simutanous small- and wide-angle X-ray scattering (SAXS/WAXS), grazing incidence small-angle X-ray (GISAXS), and grazing incidence wide-angle X-ray (GIWAXS). Follow our 12-ID-B local website for more user information. 12-ID-B covers a wide range of science from Nanomaterials and Polymers to Structural Biology and Biophysics.
|
| Research Highlights |
|
Structural basis of peptidoglycan endopeptidase regulation (2020) Most bacteria surround themselves with a cell wall, a strong meshwork consisting primarily of the polymerized aminosugar peptidoglycan (PG). PG is essential for structural maintenance of bacterial cells, and thus for viability. PG is also constantly synthesized and turned over; the latter process is mediated by PG cleavage enzymes, for example, the endopeptidases (EPs). EPs themselves are essential for growth but also promote lethal cell wall degradation after exposure to antibiotics that inhibit PG synthases (e.g., β-lactams). Thus, EPs are attractive targets for novel antibiotics and their adjuvants. However, we have a poor understanding of how these enzymes are regulated in vivo, depriving us of novel pathways for the development of such antibiotics. Here, we have solved crystal structures of the LysM/M23 family peptidase ShyA, the primary EP of the cholera pathogen Vibrio cholerae. Our data suggest that ShyA assumes two drastically different conformations: a more open form that allows for substrate binding and a closed form, which we predicted to be catalytically inactive. Mutations expected to promote the open conformation caused enhanced activity in vitro and in vivo, and these results were recapitulated in EPs from the divergent pathogens Neisseria gonorrheae and Escherichia coli. Our results suggest that LysM/M23 EPs are regulated via release of the inhibitory Domain 1 from the M23 active site, likely through conformational rearrangement in vivo.
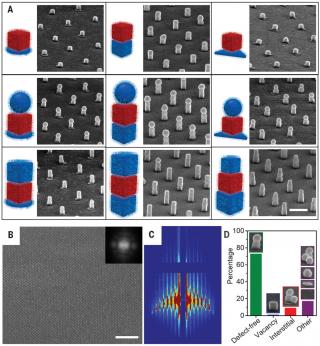
Building superlattices from individual nanoparticles via template-confined DNA (2018) DNA programmable assembly has been combined with top-down lithography to construct superlattices of discrete, reconfigurable nanoparticle architectures on a gold surface over large areas. Specifically, the assembly of individual colloidal plasmonic nanoparticles with different shapes and sizes is controlled by oligonucleotides containing “locked” nucleic acids and confined environments provided by polymer pores to yield oriented architectures that feature tunable arrangements and independently controllable distances at both nanometer- and micrometer-length scales. These structures, which would be difficult to construct by other common assembly methods, provide a platform to systematically study and control light-matter interactions in nanoparticle-based optical materials. The generality and potential of this approach are explored by identifying a broadband absorber with a solvent polarity response that allows dynamic tuning of visible light absorption.
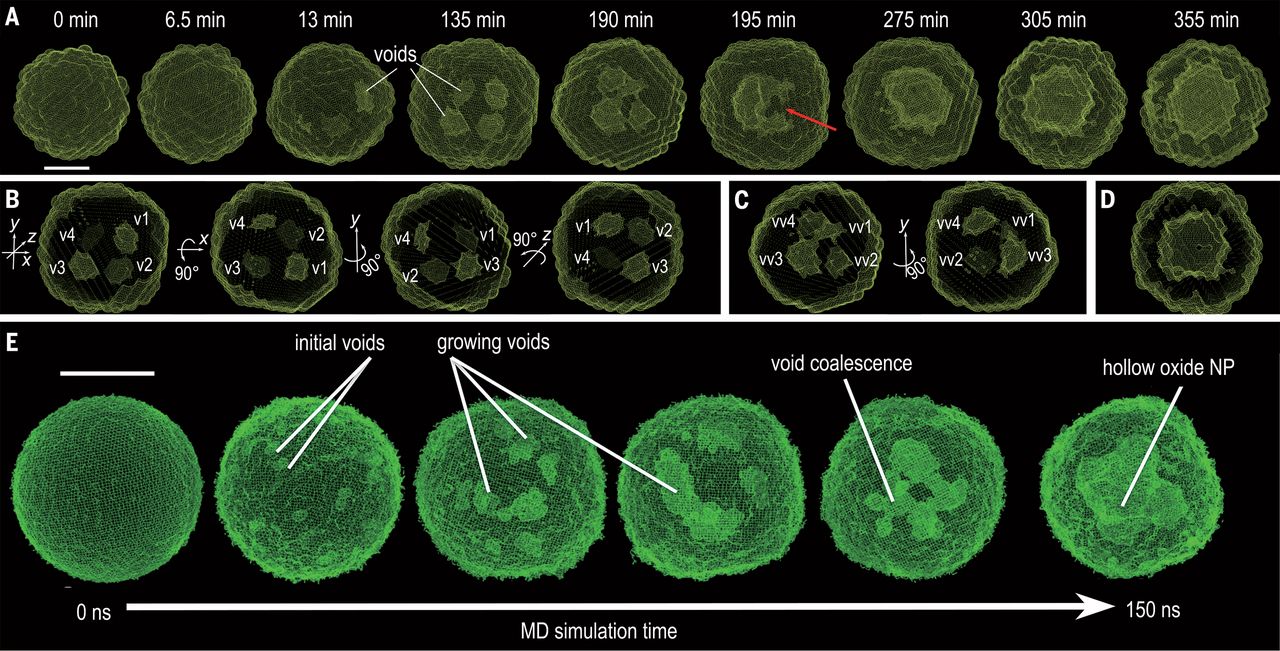
Quantitative 3D evolution of colloidal nanoparticle oxidation in solution (2017)
Real-time tracking of the three-dimensional (3D) evolution of colloidal nanoparticles in solution is essential for understanding complex mechanisms involved in nanoparticle growth and transformation. We used time-resolved small-angle and wide-angle x-ray scattering simultaneously to monitor oxidation of highly uniform colloidal iron nanoparticles, enabling the reconstruction of intermediate 3D morphologies of the nanoparticles with a spatial resolution of ~5 angstroms. The in situ observations, combined with large-scale reactive molecular dynamics simulations, reveal the details of the transformation from solid metal nanoparticles to hollow metal oxide nanoshells via a nanoscale Kirkendall process—for example, coalescence of voids as they grow and reversal of mass diffusion direction depending on crystallinity. Our results highlight the complex interplay between defect chemistry and defect dynamics in determining nanoparticle transformation and formation. |